This is the blog section. It has two categories: News and Releases.
Files in these directories will be listed in reverse chronological order.
This is the multi-page printable view of this section. Click here to print.
This is the blog section. It has two categories: News and Releases.
Files in these directories will be listed in reverse chronological order.
In June 2023, Dr. Leslie Claar and colleagues from the Allen Institute published their groundbreaking findings in the journal eLife in the article, “Cortico-thalamo-cortical interactions modulate electrically evoked EEG responses in mice”. Their approach was comprehensive: they stimulated mouse cortex while simultaneously recording with electroencephelography (EEG) and neuropixels probes, comparing brain activity during wakefulness and under isoflurane anesthesia.
In awake mice, Claar’s team observed a robust event-related potential (ERP) when they delivered pulsatile electrical stimulation to deep layers of the motor cortex (Figure 1A left). This ERP coincided with a distinct pattern of neural spiking, both locally in the motor cortex and in distant sensorimotor-related thalamic nuclei (Figure 1B left). The response followed a three-act play: initial excitation within 25 milliseconds, a period of quiescence for about 125 milliseconds, and finally, a strong rebound excitation before returning to baseline. When the mice were under anesthesia, the story changed dramatically. The ERP’s amplitude and complexity plummeted in response to the same stimulation (Figure 1A right). While the initial excitation persisted in the motor cortex, it vanished in the thalamic nuclei (Figure 1B right). The rebound excitation, along with its corresponding ERP component at around 180 milliseconds, was significantly weakened.
Based on these observations, Claar and colleagues proposed that interactions between the cortex and thalamus are responsible for the rebound excitation phase of the stimulus response. This rebound excitation, in turn, drives the second component of the ERP – a crucial signal of wakefulness.
After their experiments, Claar and her team published their data on DANDI, ensuring that it would remain publicly accessible for any future researchers who wanted to use it. The dataset and manuscript were prepared and published concurrently, which ensured that the dataset included all the paper-relevant data and metadata and curious reviewers could inspect the data directly if they so wished.
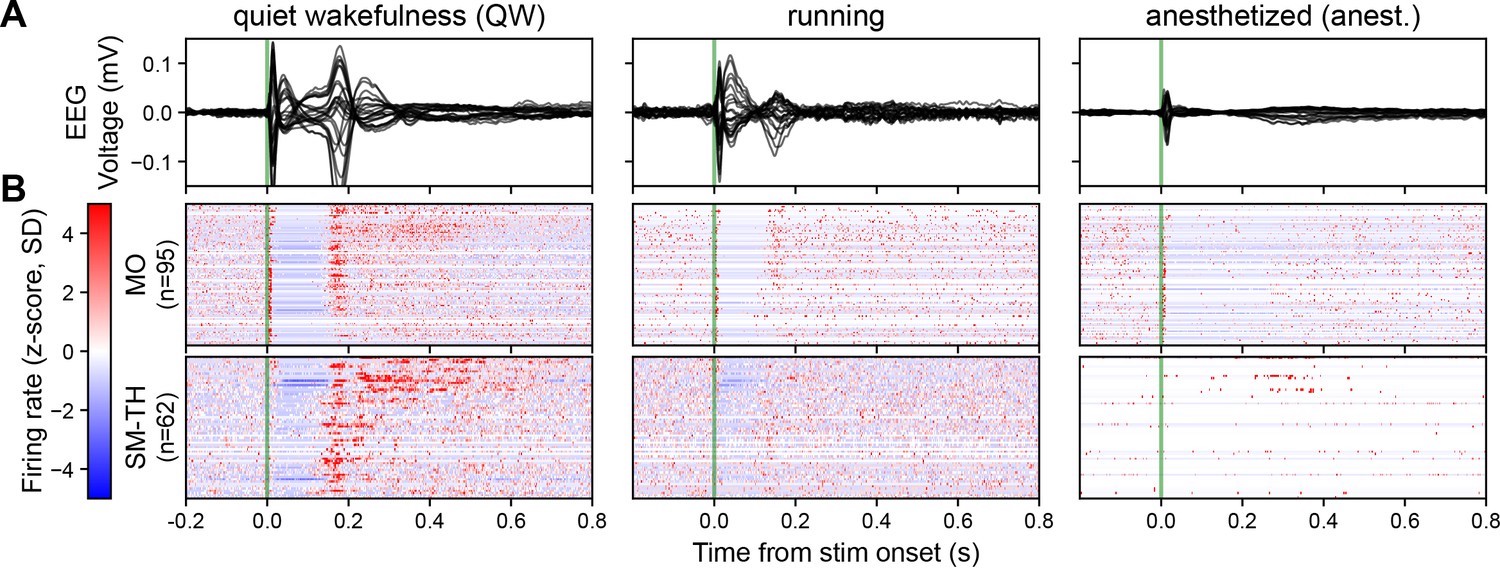
Figure 1: Brain state modulates the ERP via cortico-thalamo-cortical interactions. (A) Butterfly plot of ERPs during non-running (quiet wakefulness), running (active wakefulness), and isoflurane-anesthetized states. (B) Normalized firing rate, reported as a z-score of the average, pre-stimulus firing rate, of all RS neurons recorded by the Neuropixels probes targeting the stimulated cortex (MO) and SM-TH. Reproduced from “Cortico-thalamo-cortical interactions modulate electrically evoked EEG responses in mice”.
Just a few months later, in November 2023, Drs. Richard Burman, Paul Brodersen and colleagues at the University of Oxford wrote a paper that included reanalysis of this data. Their findings were published in the journal Neuron with the article, “Active cortical networks promote shunting fast synaptic inhibition in vivo”. They also investigated the effects of anesthesia on brain activity, but narrowed their focus to the differential function of a specific receptor subtype: GABAAR.
Burman and Brodersen’s team used a technique called in vivo gramicidin perforated patch-clamp recording, combined with optogenetic activation of GABAergic synaptic inputs. They compared the GABAA receptor equilibrium potential (EGABAAR) in mice during wakefulness and under urethane anesthesia.
Their findings were striking. In anesthetized mice, GABAA receptor responses were significantly hyperpolarizing, with an average equilibrium potential of -80.8mV. In awake mice, however, these responses could be either hyperpolarizing or depolarizing, averaging at -63.9mV – a state known as the “shunting” regime.
Burman et al. hypothesized that the higher network activity in the awake state leads to a higher concentration of intracellular chloride, which in turn depolarizes the GABAA receptor equilibrium potential. Anesthesia, by broadly suppressing neural activity, indirectly hyperpolarizes EGABAAR. They confirmed this hypothesis by locally injecting an AMPA receptor antagonist (NBQX) which suppressed network activity, and hyperpolarized EGABAAR to -78.0mV.
The Oxford researchers also hypothesized that changes in EGABAAR would lead to changes in network activity, resulting in a bi-directional relationship between the two quantities.
To test this hypothesis, Burman and Brodersen’s team turned to the data from Dandiset #000458 – the very same dataset used by Claar et al. By reanalyzing this data and manipulating a computational network model, they found that setting the EGABAAR parameter to the shunting regime (-60mV) reproduced spiking responses similar to those seen in awake mice (Figure 2 B&F left). Conversely, setting it to the hyperpolarizing regime (-80mV) mimicked the responses seen in anesthetized mice (Figure 2 B&F right). This data provides evidence for the hypothesized bidirectional relationship.
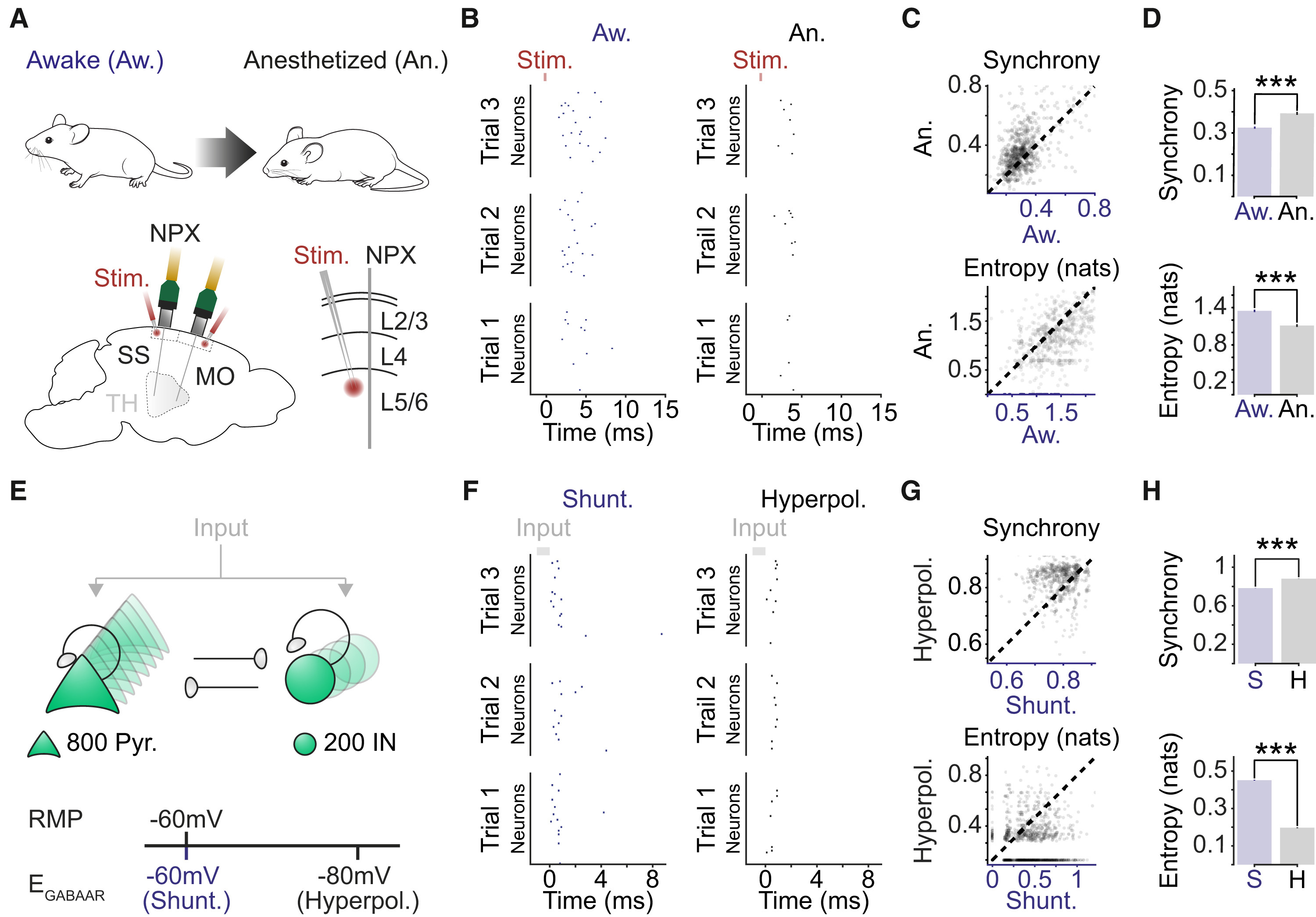
Figure 2: Shunting inhibition promotes local network desynchronization and response flexibility. (A) High-density Neuropixels (NPXs) recordings were used to compare spiking activity in the same cortical neurons under awake and anesthetized conditions. (B) Example raster plots show the spiking activity of a population of neurons in somatosensory cortex (SS). Spiking activity is shown across three stimulation trials for each condition. (C) Neuronal synchrony on a trial-to-trial basis (top) and the entropy of the peri-stimulus histogram (bottom) were calculated for each of the 662 recorded neurons under awake and anesthetized conditions (17 probe recordings in 16 mice). Each dot corresponds to a single neuron and the dashed line indicates the line of equality. (D) Neurons in the awake condition exhibited lower synchrony (top; Aw.: 0.325 ± 0.005 vs. An.: 0.393 ± 0.007, p < 0.001, paired t test) and higher entropy (bottom; Aw.: 1.347 ± 0.018 nats vs. An.: 1.111 ± 0.020 nats, p < 0.001, paired t test). (E) Schematic of network model consisting of interconnected excitatory pyramidal neurons (Pyr.) and inhibitory interneurons (IN). EGABAAR in the pyramidal neurons was adjusted relative to the RMP to create two different conditions: a shunting (Shunt.) and a hyperpolarizing (Hyperpol.) EGABAAR condition. Spiking activity was evoked by delivering brief depolarizing currents (input) of varying amplitudes to each neuron in the network. (F) Raster plots for the same population of pyramidal neurons (n = 50) in the shunting (left) and hyperpolarizing (right) EGABAAR conditions. (G) Synchrony (top) and entropy (bottom) for pyramidal neurons in the shunting and hyperpolarizing EGABAAR conditions (n = 1,000 randomly selected). Dashed line indicates the line of equality. (H) Neurons in the shunting EGABAAR condition exhibited lower synchrony (Shunt: 0.784 ± 0.001 vs. Hyperpol: 0.881 ± 0.001, p < 0.001, paired t test) and higher entropy (Shunt: 0.451 ± 0.002 nats vs. Hyperpol: 0.197 ± 0.002 nats, p < 0.001, paired t test) (n = 16,000 pyramidal neurons from the model). ∗∗∗p < 0.001. MO, motor cortex; Stim., electrical stimulation; TH, thalamus. Reproduced from “Active cortical networks promote shunting fast synaptic inhibition in vivo”.
Together, these two studies provide a rich, multi-level account of what happens to brain activity under anesthesia. They span from macroscale brain signals recorded by EEG all the way down to microscale synaptic responses from a single receptor type. The population spiking data from Dandiset #000458 serves as a crucial bridge between these levels, allowing Burman et al. to explicitly connect their work to the broader field of anesthesia research.
However, it’s important to note some key differences between the two studies. Claar et al. used isoflurane anesthesia and stimulated layers 5/6 of the cortex, while Burman et al. used urethane anesthesia and recorded from layer 2/3. Additionally, while Claar et al. found the most dramatic differences between waking and anesthetized states around 180 ms after stimulation, Burman et al. focused their reanalysis on the initial spiking response within 15 ms of stimulation.
These differences highlight the potential for further reanalysis of Dandiset #000458. Future studies could explore more similar experimental conditions, conduct a more thorough treatment of relevant response time periods, or investigate different aspects of anesthesia’s effects altogether.
For those inspired by the work of Claar, Burman and Brodersen, and their teams, the journey doesn’t end here. We encourage readers from all backgrounds to take advantage of this open science platform by publishing and reanalyzing data on DANDI. We are confident that it will be a smooth and rewarding experience, but don’t just take our word for it – listen to what Dr. Leslie Claar and Dr. Paul Brodersen have to say,
“After organizing our dataset into NWB format, validating and uploading our files was straightforward using a few simple commands from the DANDI Python Client. The tools are well-documented, which made the process of uploading and publishing quite smooth. Within a few months of publishing on DANDI, others were already interested in using our dataset.” – Dr. Leslie Claar
“The DANDI archive is a fantastic resource with great tooling and exhaustive documentation. Its command line interface makes data retrieval and synchronization as simple as possible (but not simpler!). The standardized and well-documented formats for electrophysiological and neuroimaging data ensure that anyone can quickly find their way around a new archive without requiring extensive feedback from the authors that originally collected the data. These features make the DANDI archive a treasure trove of useful and reusable data, unlike the unfortunately still commonplace unstructured data dumps on many other platforms or – worst of all – university servers.” – Dr. Paul Brodersen and Professor Colin Akerman
To get you started on your own reanalysis project, we’ve prepared a Jupyter notebook demonstrating some basic reanalysis of Dandiset #000458. This tool will help you dive right into the data, guiding you through the initial steps of exploration and analysis. We have also prepared a Neurosift walkthrough which demonstrates how to use Neurosift to quickly and easily explore the NWB files in this dataset without even leaving the browser.
By engaging with this open science platform, you become part of a larger community of researchers who are building on each other’s work and driving the field forward. In the world of open science, collaboration and curiosity drive discovery—so why not see where the data takes you?
Date/Time: April 21 - 22, 2022. 9am - 6pm ET
Online: Link will be sent to registered participants.
If you are interested in participating please fill in this form to be sent announcements: Register here
The events will run 9 - 6 ET to accommodate multiple time zones. Participants are welcome to join at any time.
DANDI+Microscopy Starter Kit: We will generate a set Starter Kit that makes it easy for users to navigate these and other Dandisets, including easier visualization of individual files.
Dandiset Abstraction Layer:
Unconference session (11.30 - 12.30 ET)
Visualization/Annotation:
Registration:
Unconference session (11.30 - 12.30 ET)
Please feel free to comment on this community doc.
Neurophysiology experiments often include natural videos (such as behaving animals), which need to be stored with the neurophysiological recordings in order to ensure maximal reusability of the data. These videos are commonly stored with lossy compression (e.g. h264 in an .mp4 file), which allows them to achieve very high compression ratios. It is possible to read these videos frame-by-frame, and store them in HDF5, but since HDF5 is not able to access popular video codecs like h264, the volume of the video in the NWB file is much larger (even when using the available compression algorithms like GZIP). NWB has an option to avoid storing these altogether by linking to these external video files using a relative path to that file on disk. This relative path is stored in the ImageSeries neurodata_type storing it as an attribute of a string dtype. We also need to publish these video linked NWB files in an archive (e.g. in DANDI). For DANDI, which renames and reorganizes the the NWB files, this requires not only uploading the video file on the archive but also changing the path attribute of the ImageSeries to reflect the new file names.
To implement this, we have created a formal naming convention for these video files relative to the NWB files’ path. In addition, these video files are also placed in a specific folder structure relative to the new location of the NWB file during the dandi organize call.
Internally the steps are as follows:
external_file attribute in the NWB files.Note: this solution is specifically for natural videos like those of behaving animals. There are other types of image sequences like image stacks from optical physiology, which do not use codecs like h264; these types of videos can be copied into an HDF5 file.
├── nwbfiles
│ ├── test1_0_0.nwb
│ └── test1_1_1.nwb
└── video_files
├── test1_0.avi
├── test1_1.avi
├── test2_0.avi
└── test2_1.avi
With the path attribute as: image_series.external_files=["../video_files/test1_0.avi", "../video_files/test1_1.avi"]
dandi organizeThe renaming pattern is as follows /<nwbfile_name>/{ImageSeries UUID}_external_file_{number}.mp4.
This UUID is that assigned to the ImageSeries datatype when its created. Thus its possible to lookup a video file linked to an NWB file and vice versa.
└── dandi_organized
├── sub-mouse0
│ ├── sub-mouse0_ses-sessionid0_image
│ │ ├── 933f8cf6-9e4b-405f-8cad-cc031d1fafc9_external_file_0.avi
│ │ └── 933f8cf6-9e4b-405f-8cad-cc031d1fafc9_external_file_1.avi
│ └── sub-mouse0_ses-sessionid0_image.nwb
└── sub-mouse1
├── sub-mouse1_ses-sessionid1_image
│ ├── 03137112-9d42-46b6-9046-45bc9aa7eb5e_external_file_0.avi
│ └── 03137112-9d42-46b6-9046-45bc9aa7eb5e_external_file_1.avi
└── sub-mouse1_ses-sessionid1_image.nwb
With the renamed path attribute as
image_series.external_files=
["sub-mouse0_ses-sessionid0_image/933f8cf6-9e4b-405f-8cad-cc031d1fafc9_external_file_0.avi",
"sub-mouse0_ses-sessionid0_image/933f8cf6-9e4b-405f-8cad-cc031d1fafc9_external_file_1.avi"]
cd dandi_organized
dandi download "https://gui-staging.dandiarchive.org/#/dandiset/101391/draft"
cd dandi_organized
dandi organize -f "copy" --update-external-file-paths --media-files-mode "copy" "/nwbfiles"
–modify-external-file-fields option is a flag.
If active, the organise operation modifies the external_file field of an ImageSeries that holds the local location of an associated video file. It changes the value to the new name as per the convention above.
If no external_file field found in all nwb files, but this option is active, then it logs a warning.
If any NWB file’s ImageSeries has a external_file, but this option is not specified, then it raises a ValueError to avoid breaking the link.
–media-files-mode can be any of copy/move/symlink/hardlink.
This can only be specified if the –modify-external-file-fields flag is True. This is an optional argument, if not specified it defaults to “symlink”: an efficient way to deal with possibly large video files.
dandi validate
dandi upload -i dandi-staging "/dandi_organized"
Example dandiset here
This dataset can then be downloaded using:
mkdir dandi_download
cd dandi_download
dandi download "https://gui-staging.dandiarchive.org/#/dandiset/101391/draft"
The folder will contain all the video files along with the dandi metadata .yml and .nwb files.
Click to view a recording of the workshop
DANDI is a US BRAIN Initiative supported data archive for publishing and sharing neurophysiology data including intracellular and extracellular electrophysiology, optophysiology, and behavioral time-series, and images from immunostaining experiments. For example, data from experimental techniques like patch clamps, silicon probes, and calcium imaging can be published on DANDI. So can data from lightsheet microscopy experiments when combined with associated MRI data.
DANDI allows scientists to publish “dandisets,” collections of data from neuroscience experiments that are often associated with specific papers or projects. DANDI is built to scale with the growing data engineering needs of the community and can easily receive and publish TBs of data for free supported by the AWS public dataset program. It also uses best practices in metadata curation using the DANDI schema and allows users to mint DOIs using Datacite for publishing dandisets.
For neurophysiology data, DANDI supports the Neurodata Without Borders (NWB) standard. The NWB standard ensures that data is shared with the necessary metadata for reanalysis, allows for analyses that aggregate data across dandisets, provides a common platform for building analysis and visualization tools for published data, and allows DANDI to easily extract metadata to provide powerful detailed search features. For MRI and related data, DANDI has been leveraging the BIDS standards and working with other groups to extend support to microscopy and electrophysiology.
With DANDI Hub, DANDI is more than just an archive, but in fact an entire cloud-based collaborative platform, available for free to all DANDI users! DANDI Hub is a JupyterHub with access to high performance multi-core compute nodes and high-bandwidth access to all data hosted on DANDI. DANDI Hub provides many different workflows to interact with your data, including Python, R, and Julia Jupyter notebooks, a terminal, and even a Linux desktop view, all deployed for free and on demand in the cloud. The hub can be used to share code that reproduces key figures from papers, or to interactively explore any dandiset.
You can navigate all data conveniently using the Web app, or for more control we have several software tools for interacting with DANDI. The DANDI Python library and command line interface allows for performant upload, download, and querying. The DANDI REST API can also be used to interface with web applications. Also, all dandisets are made available using Datalad, which provides convenient data access across different systems.
Upload your data today! See our user guide for details. If you have any questions, please use our help desk and we will be happy to assist.